He has been selected as the “Highly Cited Researchers” for five consecutive years
(Cross-Field – 2018, 2022; Plant and Animal Science – 2019, 2020, 2021).

Laboratory of Crop Evolution, Kyoto University




Genomic Dynamics Underlying the Plastic Hermaphroditism in Plants: https://www.ige.tohoku.ac.jp/prg/flower/ (in japanese)
Chia, K.-S., Kourelis, J., Vickers, M., Sakai, T., Kamoun, S., & Carella, P. (2022). The N-terminal executioner domains of NLR immune receptors are functionally conserved across major plant lineages. BioRxiv, 2022.10.19.512840. https://doi.org/10.1101/2022.10.19.512840 de la Concepcion, J. C., Fujisaki, K., Bentham, A. R., Cruz Mireles, N., Sanchez de Medina Hernandez, V., Shimizu, M., Lawson, D. M., Kamoun, S., Terauchi, R. & Banfield, M. J. (2022). A blast fungus zinc-finger fold effector binds to a hydrophobic pocket in host Exo70 proteins to modulate immune recognition in rice. Proceedings of the National Academy of Sciences, 119(43), e2210559119. https://doi.org/10.1073/pnas.2210559119
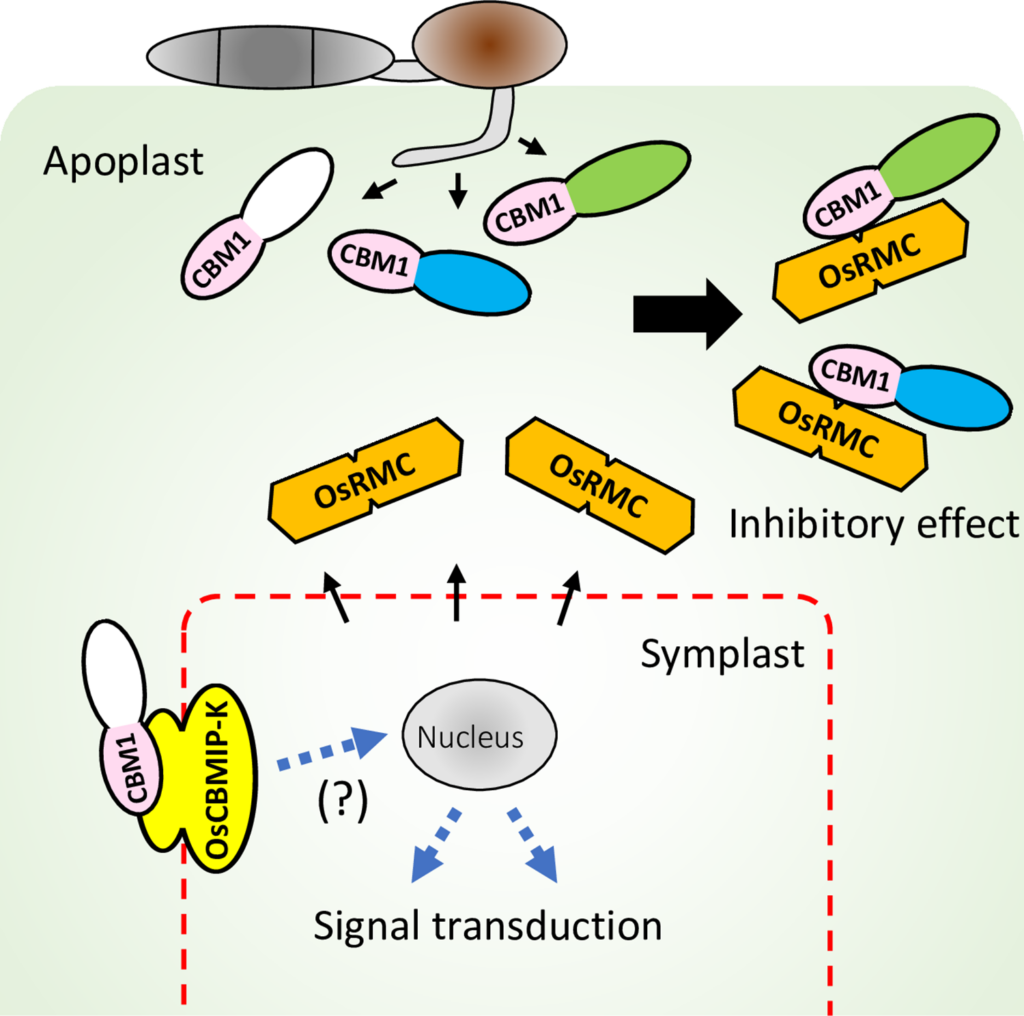
Takeda, T., Takahashi, M., Shimizu, M., Sugihara, Y., Yamashita, T., Saitoh, H., Fujisaki, K., Ishikawa, K., Utsushi, H., Kanzaki, E., Sakamoto, Y., Abe, A., & Terauchi, R. (2022). Rice apoplastic CBM1-interacting protein counters blast pathogen invasion by binding conserved carbohydrate binding module 1 motif of fungal proteins. PLOS Pathogens, 18(9), e1010792-. https://doi.org/10.1371/journal.ppat.1010792
When infecting plants, fungal pathogens secrete cell wall-degrading enzymes (CWDEs) that break down cellulose and hemicellulose, the primary components of plant cell walls. Some fungal CWDEs contain a unique domain, named the carbohydrate binding module (CBM), that facilitates their access to polysaccharides. However, little is known about how plants counteract pathogen degradation of their cell walls. Here, we show that the rice cysteine-rich repeat secretion protein OsRMC binds to and inhibits xylanase MoCel10A of the blast fungus pathogen Magnaporthe oryzae, interfering with its access to the rice cell wall and degradation of rice xylan. We found binding of OsRMC to various CBM1-containing enzymes, suggesting that it has a general role in inhibiting the action of CBM1. OsRMC is localized to the apoplast, and its expression is strongly induced in leaves infected with M. oryzae. Remarkably, knockdown and overexpression of OsRMC reduced and enhanced rice defense against M. oryzae, respectively, demonstrating that inhibition of CBM1-containing fungal enzymes by OsRMC is crucial for rice defense. We also identified additional CBM-interacting proteins (CBMIPs) from Arabidopsis thaliana and Setaria italica, indicating that a wide range of plants counteract pathogens through this mechanism.
Plants have evolved various activity-inhibiting proteins as a defense against fungal cell wall-degrading enzymes (CWDEs), but how plants counteract the function of fungal enzymes containing carbohydrate binding modules (CBMs) remains unknown. Here, we demonstrate that OsRMC, a member of the cysteine-rich repeat secretion protein family, interacts with fungal CBM1. OsRMC binding to CBM1 of a blast fungal xylanase blocks access to cellulose, resulting in the inhibition of xylanase enzymatic activity. Our study provides significant insights into plant countermeasures against CWDEs in the apoplastic space during plant-fungal pathogen interactions. It also reveals a molecular function of the DUF26 domain widely distributed in plant proteins.