Sugihara, Y., Abe, Y., Takagi, H., Abe, A., Shimizu, M., Ito, K., Kanzaki, E., Oikawa, K., Kourelis, J., Langner, T., Win, J., Białas, A., Lüdke, D., Contreras, M. P., Chuma, I., Saitoh, H., Kobayashi, M., Zheng, S., Tosa, Y., Banfield, M., Kamoun, S., Terauchi, R., & Fujisaki, K. (2023). Disentangling the complex gene interaction networks between rice and the blast fungus identifies a new pathogen effector. PLOS Biology, 21(1), e3001945-. https://doi.org/10.1371/journal.pbio.3001945 Kovi, B., Sakai, T., Abe, A., Kanzaki, E., Terauchi, R., & Shimizu, M. (2023). Isolation of Pikps, an allele of Pik, from the aus rice cultivar Shoni. Genes & Genetic Systems, advpub. https://doi.org/10.1266/ggs.22-00002
An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana
This paper has been published in which Dr. Adachi is the corresponding author.
Adachi, H., Sakai, T., Harant, A., Pai, H., Honda, K., Toghani, A., Claeys, J., Duggan, C., Bozkurt, T. O., Wu, C., & Kamoun, S. (2023). An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana. PLOS Genetics, 19(1), e1010500-. https://doi.org/10.1371/journal.pgen.1010500
Abstract
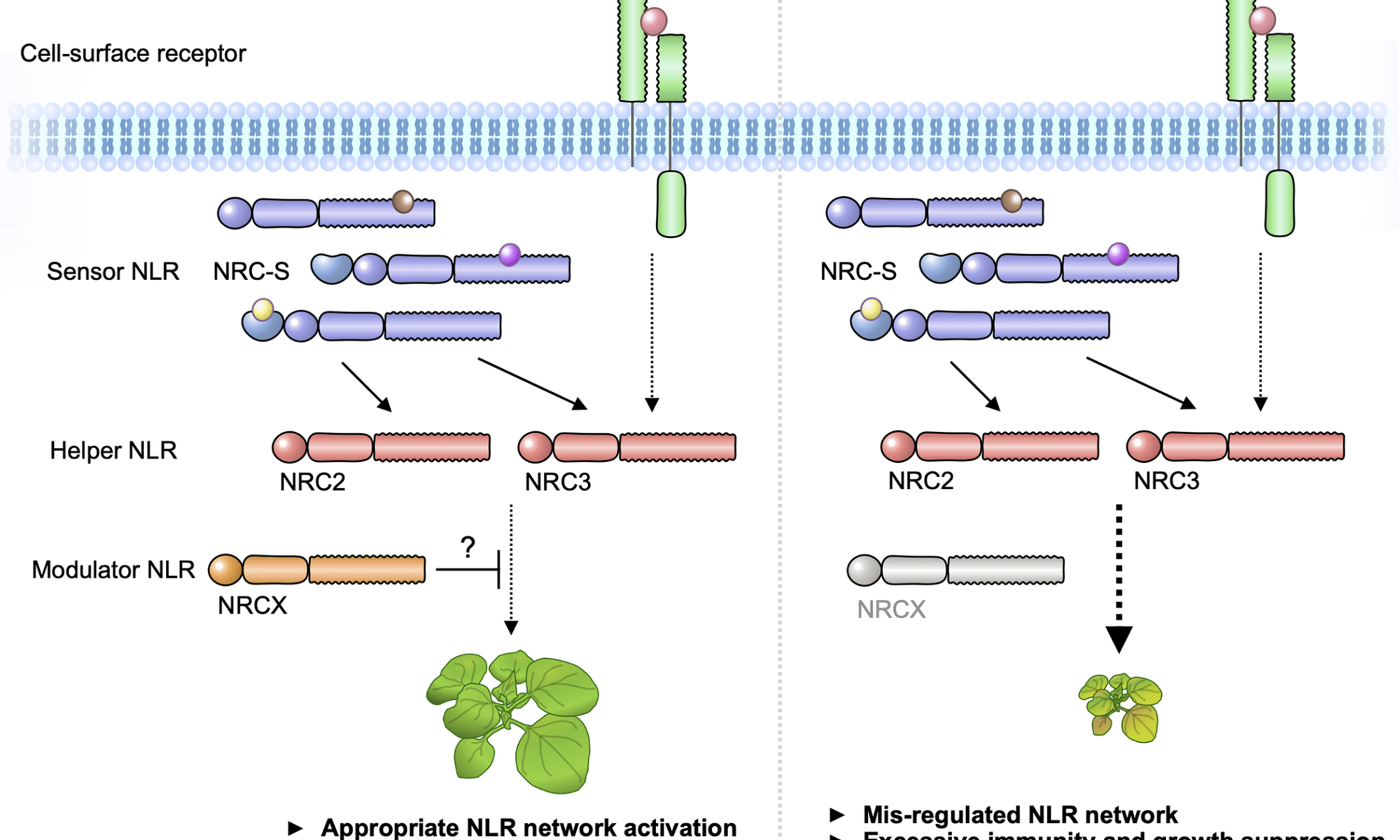
The NRC immune receptor network has evolved in asterid plants from a pair of linked genes into a genetically dispersed and phylogenetically structured network of sensor and helper NLR (nucleotide-binding domain and leucine-rich repeat-containing) proteins. In some species, such as the model plant Nicotiana benthamiana and other Solanaceae, the NRC (NLR-REQUIRED FOR CELL DEATH) network forms up to half of the NLRome, and NRCs are scattered throughout the genome in gene clusters of varying complexities. Here, we describe NRCX, an atypical member of the NRC family that lacks canonical features of these NLR helper proteins, such as a functional N-terminal MADA motif and the capacity to trigger autoimmunity. In contrast to other NRCs, systemic gene silencing of NRCX in N. benthamiana markedly impairs plant growth resulting in a dwarf phenotype. Remarkably, dwarfism of NRCX silenced plants is partially dependent on NRCX paralogs NRC2 and NRC3, but not NRC4. Despite its negative impact on plant growth when silenced systemically, spot gene silencing of NRCX in mature N. benthamiana leaves doesn’t result in visible cell death phenotypes. However, alteration of NRCX expression modulates the hypersensitive response mediated by NRC2 and NRC3 in a manner consistent with a negative role for NRCX in the NRC network. We conclude that NRCX is an atypical member of the NRC network that has evolved to contribute to the homeostasis of this genetically unlinked NLR network.
Author summary
Plants have an effective immune system to fight off diverse pathogens such as fungi, oomycetes, bacteria, viruses, nematodes and insects. In the first layer of their immune system, receptor proteins act to detect pathogens and activate the defense response. Plant genomes encode very large and diverse repertoires of immune receptors, some of which function in pairs or as complex receptor networks. However, the immune system can come at a cost for plants and inappropriate receptor activation results in growth suppression and autoimmunity. Here, we show that an atypical immune receptor gene functions as a modulator of the immune receptor network. This type of receptor gene evolved to maintain homeostasis of the immune system and balance fitness trade-offs between growth and immunity. Further understanding how plants regulate their immune receptor system should help guide breeding disease resistant crops with limited fitness penalties.
We propose that “Modulator NLR” contributes to NLR immune receptor network homeostasis during plant growth. A modulator NRCX has a similar sequence signature with helper MADA-CC-NLRs, but unlike helpers, NRCX lacks the functional MADA motif to execute cell response. NRCX modulates the NRC2/NRC3 subnetwork composed of multiple sensor NLRs and cell-surface receptor (left). Loss of function of NRCX leads to the enhanced hypersensitive response and dwarfism in N. benthamiana plants (right).
Updated our publications list.
Adachi, H., Sakai, T., Harant, A., Pai, H., Honda, K., Toghani, A., Claeys, J., Duggan, C., Bozkurt, T. O., Wu, C., & Kamoun, S. (2023). An atypical NLR protein modulates the NRC immune receptor network in Nicotiana benthamiana. PLOS Genetics, 19(1), e1010500-. https://doi.org/10.1371/journal.pgen.1010500 Kourelis, J., Contreras, M. P., Harant, A., Pai, H., Lüdke, D., Adachi, H., Derevnina, L., Wu, C.-H., & Kamoun, S. (2022). The helper NLR immune protein NRC3 mediates the hypersensitive cell death caused by the cell-surface receptor Cf-4. PLOS Genetics, 18(9), e1010414-. https://doi.org/10.1371/journal.pgen.1010414
Isolation of Pikps, an allele of Pik, from the aus rice cultivar Shoni
New paper from our laboratory
Kovi, B., Sakai, T., Abe, A., Kanzaki, E., Terauchi, R., & Shimizu, M. (2023). Isolation of Pikps, an allele of Pik, from the aus rice cultivar Shoni. Genes & Genetic Systems, advpub. https://doi.org/10.1266/ggs.22-00002
Abstract
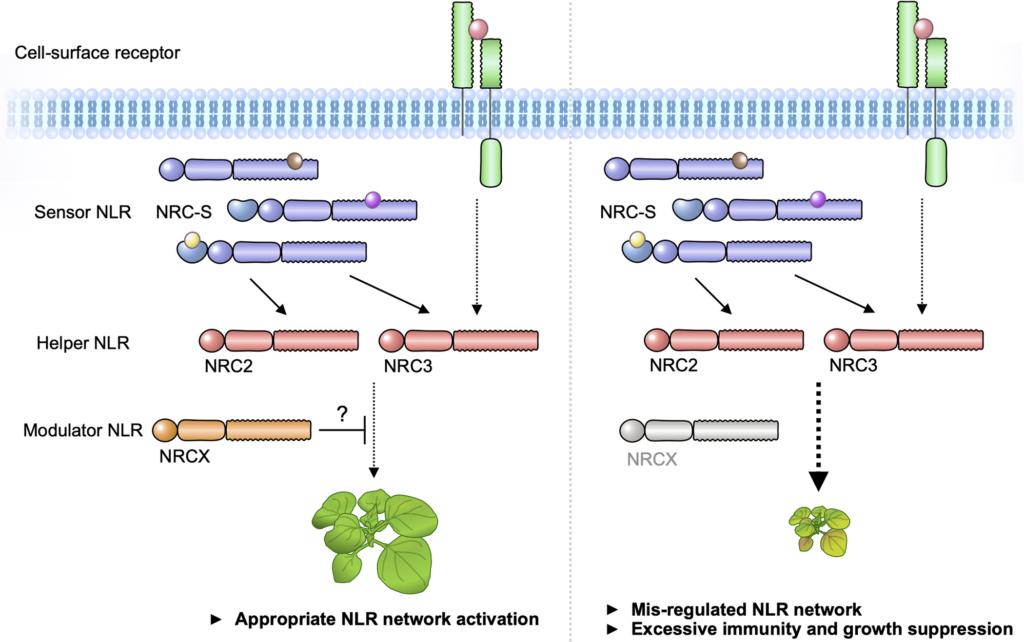
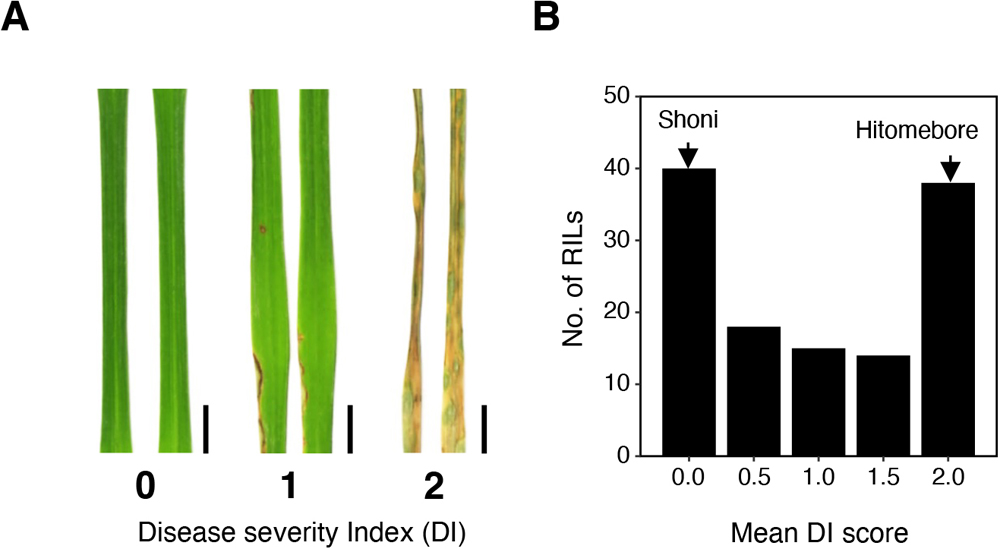
Blast disease caused by the filamentous fungus Pyricularia oryzae (syn. Magnaporthe oryzae) is one of the most destructive diseases of rice (Oryza sativa L.) around the globe. An aus cultivar, Shoni, showed resistance against at least four Japanese P. oryzae isolates. To understand Shoni’s resistance against the P. oryzae isolate Naga69-150, genetic analysis was carried out using recombinant inbred lines developed by a cross between Shoni and the japonica cultivar Hitomebore, which is susceptible to Naga69-150. The result indicated that the resistance was controlled by a single locus, which was named Pi-Shoni. A QTL analysis identified Pi-Shoni as being located in the telomeric region of chromosome 11. A candidate gene approach in the region indicated that Pi-Shoni corresponds to the previously cloned Pik locus, and we named this allele Pikps. Loss of gene function mediated by RNA interference demonstrated that a head-to-head-orientated pair of NBS-LRR receptor genes (Pikps-1 and Pikps-2) are required for the Pikps-mediated resistance. Amino acid sequence comparison showed that Pikps-1 is 99% identical to Pikp-1, while Pikps-2 is identical to Pikp-2. Pikps-1 had one amino acid substitution (Pro351Ser) in the NBS domain as compared to Pikp-1. The recognition specificity of Pikps against known AVR-Pik alleles is identical to that of Pikp.
(A) The disease severity index (DI) was employed in evaluating phenotypes of RILs after spray inoculation of the fungus. DI = 0, no symptoms; DI = 1, 0 to 20% infected leaf area; DI = 2, over 20% infected leaf area. Representative leaves for each category are shown. Scale bars, 0.5 cm. (B) Frequency distribution of the DI for 125 RILs derived from a cross between Hitomebore and Shoni. Arrows indicate approximate value obtained for the parental (Hitomebore and Shoni) lines. The DI score of each RIL is represented by the average value of two technical replications.
(A) QTL analysis of DI scores obtained from the 125 RILs. The dashed line indicates the significance threshold (-log10(P) > 3.36). (B) Comparative genomic mapping of the 11 NLR genes in Nipponbare within the scaffold bctg00000014 of the Shoni genome assembly. The black arrows indicate NLR genes. Pikps-1 and Pikps-2 correspond to LOC_Os11g46200 and LOC_Os11g46210, respectively
Prof. Terauchi was selected to Clarivate’s “Highly Cited Researchers 2022”

He has been selected as the “Highly Cited Researchers” for five consecutive years
(Cross-Field – 2018, 2022; Plant and Animal Science – 2019, 2020, 2021).